Care, Health and Reasoning Machines
The CHARM laboratory is led by Jeremy Weiss. We develop probabilistic machine learning algorithms and deploy them in healthcare settings at CMU and Pittsburgh. The Heinz College at CMU holds expertise in analytics-driven health policy, excellence in Computer Science and Machine Learning, and close ties to our healthcare industry partners and innovation community.
Prospective PhD students: consider applying to Heinz College, the Joint PhD Program in Machine Learning and Public Policy, or the University of Pittsburgh PhD Program in Biomedical Informatics.
Lab members:
- Wenbin Zhang, Heinz postdoctoral fellow
- Cheng Cheng, Heinz MLPP PhD student
- Xuejian Wang, Heinz PhD student
- Shahriar Noroozidadeh, Heinz PhD student
Former lab members:
- Aishwarya Jadhav
- Beiming Zhang
- Veronica Rosas Wiscovitch
- David Ramirez Gomez
- Yoonjung Kim
News
TL-Lite: temporal visualization and learning for clinical forecasting, accepted at NeurIPS ML4H 2020.
Harmonic Mean Point Processes: an Approach to Proportional Rate Error Minimization Across Time for Obtundation Prediction is accepted as an extended abstract to the NeurIPS Workshop on Machine Learning for Health, Vancouver, 2019.
Hypersphere clustering to characterize healthcare providers using prescriptions and procedures from Medicare claims data is accepted to the American Medical Informatics Association Annual Symposium, Washington D.C., 2019.
Derivation, Validation and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis is accepted to the Journal of the American Medical Association (JAMA), 2019.
Evaluation of Machine-Learning Algorithms for Predicting Opioid Overdose Risk Among Medicare Beneficiaries With Opioid Prescriptions is accepted to the Journal of the American Medical Association (JAMA) Network Open, 2019.
Machine learning assisted discovery of novel predictive lab test using electronic health record data is accepted to the American Medical Informatics Association Informatics Summit, San Francisco, 2019.
Bicluster phenotyping of healthcare providers, procedures, and prescriptions at scale with deep learning is accepted to the Joint Workshop on Artificial Intelligence in Healthcare, Stockholm 2018.
Survival-supervised topic modeling with anchor words: characterizing pancreatitis outcomes accepted to NIPS workshop of Machine Learning for Health, 2017.
Piecewise-constant parametric approximations for survival learning is accepted to Machine Learning for Healthcare 2017.
Motivation
Why EHRs: Electronic health records (EHRs) document over 80% of medical encounters, a 7-fold increase from one decade ago, as a result of government subsidies through the HITECH Act. Using EHR data, we can now directly conduct clinical analyses to improve health outcomes, less the high cost of conducting longitudinal studies. Alongside EHR data, device and -omic data are coming online that will require scalable and integrated methods to facilitate evidence-based personalized predictions and recommendations for patients.
Call for machine learning: However, EHR data are messy. Structured data--a fraction of data contained in EHRs--come as databases, not fixed-length feature vectors. Medicine changes over time, in prevalence, treatment, and best practices. What an EHR stores also changes over time.
Conclusions from data that guide health policy will require machine learning and statistical approaches that account for such characteristics.
However, with machine learning we can understand EHR data better than we ever have before. We cast our analyses under the terms characterization, prediction, and intervention.
Ongoing projects:
Clustering at scale
Trust is a key issue for predictions from machine learning models. Personalization is a key idea for tailoring care processes. One way to assess quality of tailored prediction and engender trust is through subgroup analysis. We are developing high dimensional clustering frameworks that provide subgroup uncertainty characteristics.
k-year risk of disease y
We can flexibly automate the process of constructing a longitudinal clinical study from EHR data. Most observational studies can be constructed using sorting, filtering, and matching operations. We are working to operationalize the use of EHR data across diseases and time.
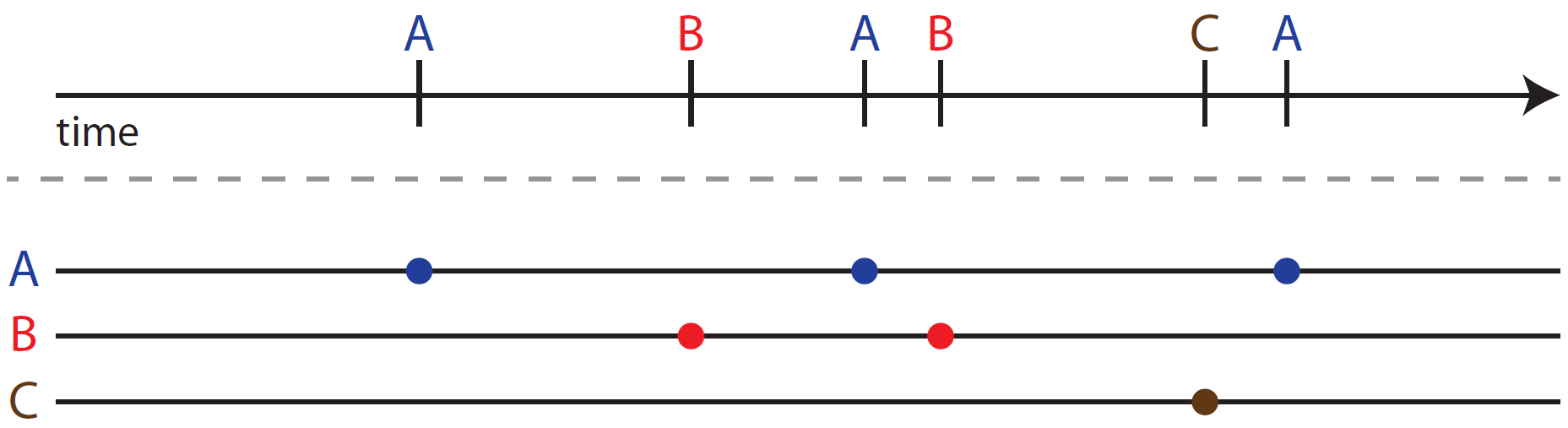
Timelines as random and regular process mixtures
E.g. a patient with diabetes has random events (like diabetic ketoacidosis, diabetic retinopathy, etc.) mixed with regular events (blood sugar measurements, eye and foot exams, A1C measurements). Modeling mixtures of random and probabilistically regular events requires machine learning model development.
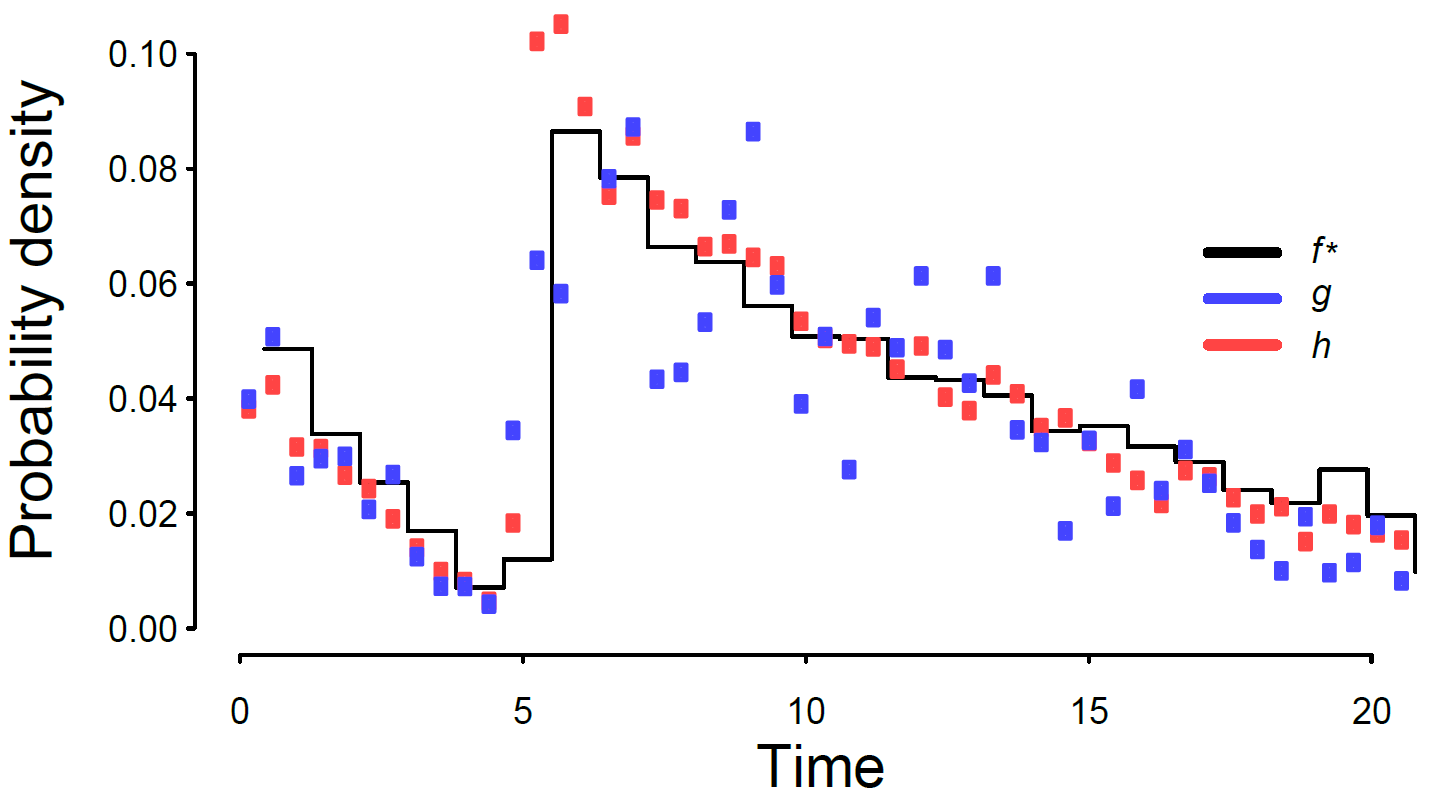
Constrained machine learning for inference
Inference is the primary limiting factor in scaling all sorts of machine learning models to big data, and many such problems have been shown to be NP-hard. In the learning-for-inference subfield, Weiss et al., AAAI 2015 (pdf) showed how approximate inference could be markedly improved by using predictions from a constrained machine learning setup. We are analyzing this problem in greater detail.
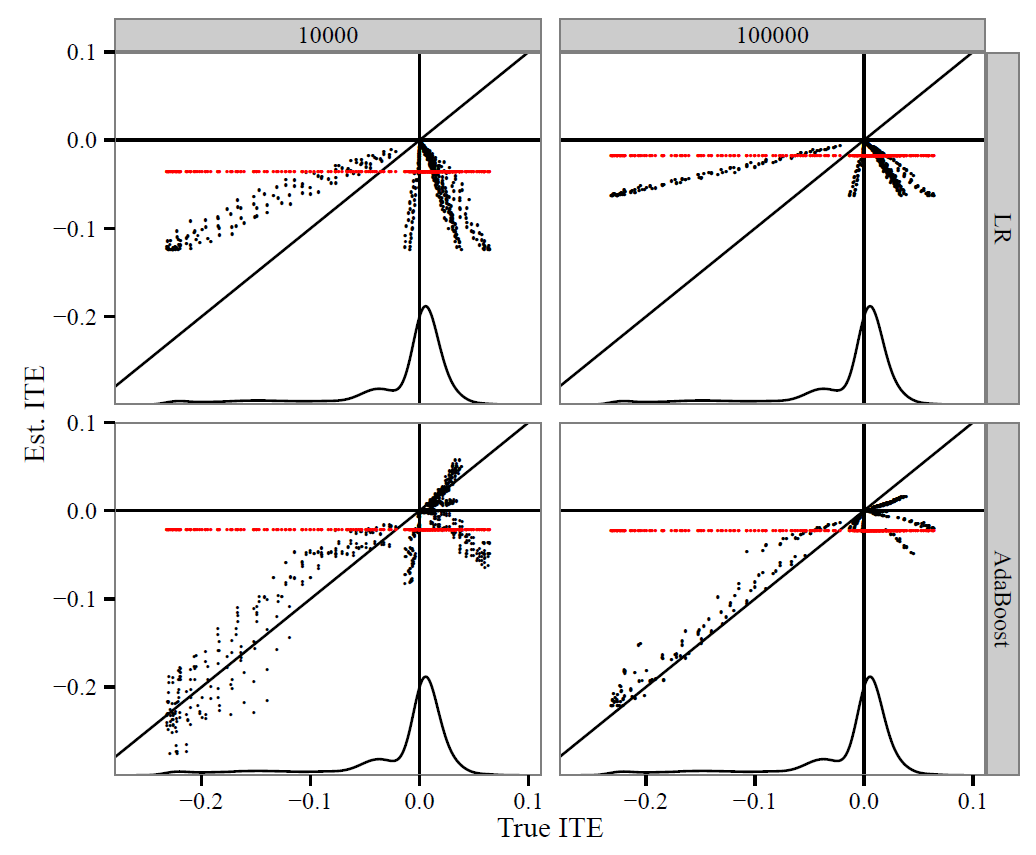
Individualized treatment effects (ITE) from randomized, semi-randomized and observational data
Randomized control trials (RCTs) aka A-B testing model the average treatment effect. To avoid heterogeneity, trials narrow their inclusion criteria, but then want to make general conclusions about the value of their therapy. Observational studies have plentiful data in populations with heterogeneity yet must make strong assumptions when used to model cause and effect. Understanding methods to estimate the individualized treatment effect are crucial for making personalized causal claims. We believe the use of semi-randomized data can aid us and are developing ITE estimators from such data.